Whole Slide Image export¶
There are videos detailing the whole slide image export process from start to finish at the end of this page. Below is a breakdown of each panel and button used in the examples.
Tissue export window¶
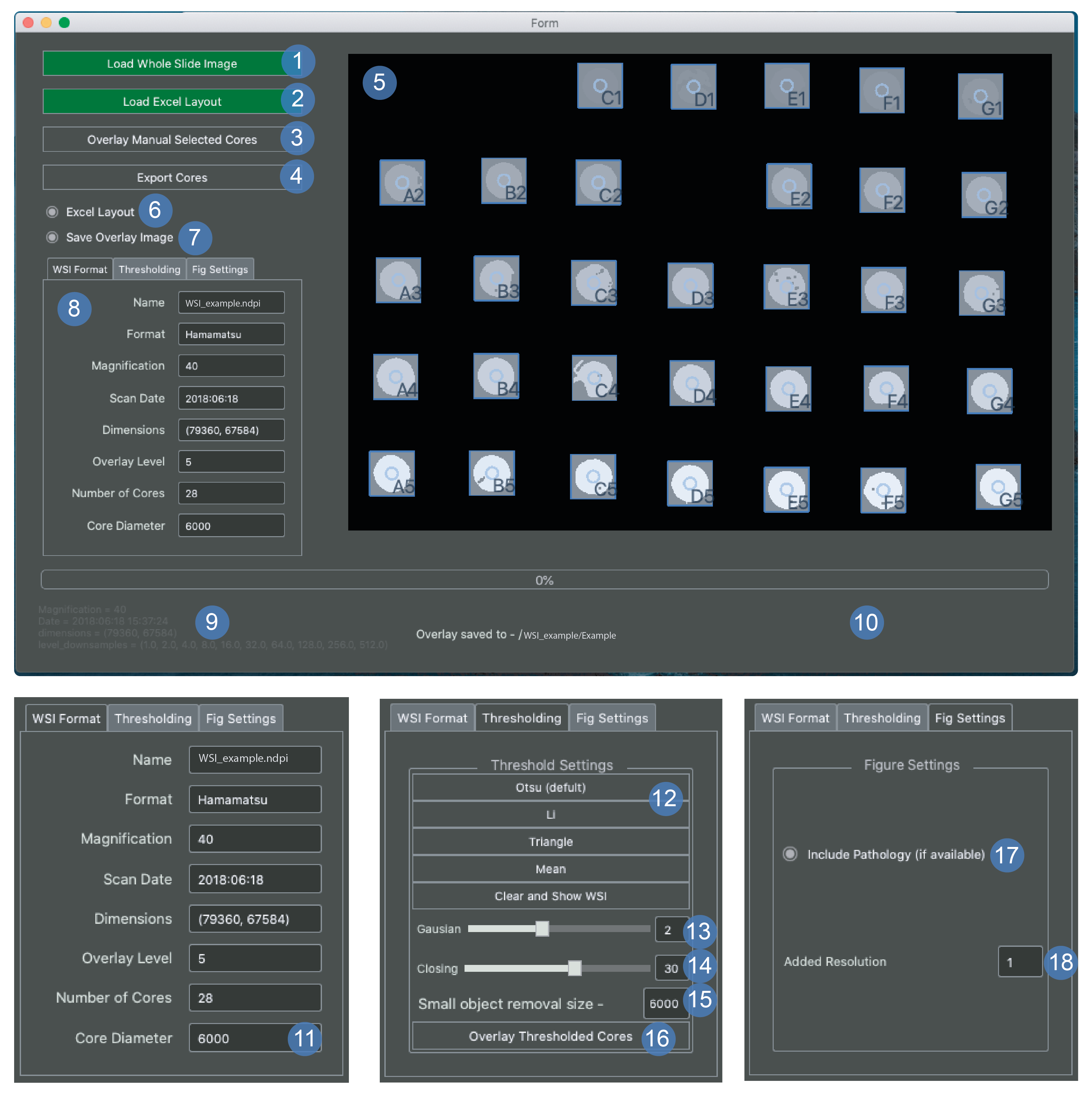
The tissue export window is for exporting png images from whole slide files using a map of the array that is provided by the user - see array map example.
Load WSI prompts a dialog box for the user to select a NDPI or SVS file.
Load Excel this is a prompt to load the excel map.
If the excel is named the same as the array and in the same directory,it will be loaded automatically. Otherwise this button can be used to load a different location.
If this has happened then the button will turn green.
This can also be used to reload the excel map at any point if the map has been changed by the user.
Overlay of manually selected cores.
This is applied after a user selection has been applied in panel number 5.
It can also be used after the cores have been moved to update the window.
Export cores should only be executed the user is satisfied that the cores are in the correct configuration.
Export core command will disable user input and export the core png images to the WSI directory.
Display window.
Double click to add core.
Spacebar to remove last point.
You can drag pre-applied cores before or after applying the bounding boxes.
Excel layout the recommended way to index cores is with the row names as the number and the column names as the letter (A6 col A row 6) if you want to reverse this (A6 row A col 6) then uncheck the box when the window opens.
Save overlay image determines whether or not to automatically save the image in the viewer (5).
Tab viewer window is further explained in points 11-18.
Image metadata for the WSI file.
Progress update panel is where progress updates will appear.
Core diameter is currently set to 6000 pixels which is similar bounding box size as a 1mm diameter tissue core.
This value can be chosen before image export and you can see what the new selection looks like by selecting the overlay cores button (3).
Thresholding options.
Choose the best threshold then proceed to 13.
Gaussian blur shows the sigma value applied to the gaussian which must be applied after 12.
Closing should be applied after 13 and stands for binary morphological closing.
higher slider values will lead to a more closed mask.
Removal of small objects from the mask gives the value of the minimum size to be removed.
If you have very small tissue cores then this will need to be reduced.
This applies the changes added in steps 12-15 and overlays the core images and labels.
These are the settings for the first figure that is exported.
This option will add a bounding box to the figure to denote the pathology of the core as either red or green.
This will only work if the first tab of the xlsx file with the array contains a map indexed with N and T for normal and tumour, respectively.
Please see the array map example for more indexing pathology details.
If you increase this number, then it will increase the level that the images are taken from in the WSI.
Increasing this will exponentially slow the program down so use sparingly.